- Startseite
- Forschung
- AG Neuroimaging & Neuroengineering
- AG Altern & Demenz
- AG Bewegungsstörungen & Kognition
- AG Bewegungsstörungen & Tiefe Hirnstimulation
- AG Epileptologie
- AG Experimentelle Neuroimmunologie
- AG Klinische Neuroimmunologie
- AG Kognitive Neurologie
- AG Network Plasticity
- AG Neurale Stammzellen
- AG Neuroimaging & Neuroengineering
- AG Neuroinflammation & neuromuskuläre Erkrankungen
- AG Neuromodulation & Neurorehabilitation
- AG Neuroonkologie
- AG Neuropsychologie
- AG Prodromalstadien und Schlaf bei Bewegungsstörungen
- AG Sensomotorische Adaptation
- AG Zerebrovaskuläre Erkrankungen
- Köln-Jülich Biobank
AG Neuroimaging und Neuroengineering des experimentellen Schlaganfalls
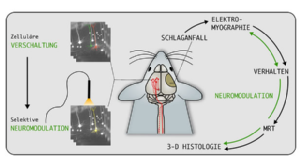
Wir untersuchen die zellulären Grundlagen von Netzwerkveränderung, die zu Störungen der Motorik nach einem experimentellen Schlaganfall führen. Ein Schlaganfall ist die häufigste Ursache für eine Langzeitbehinderung, wobei motorische Defizite eine große Rolle spielen, die zu alltagsrelevanten Einschränkungen für die Patienten führen. Für diese chronische Phase nach einem Schlaganfall gibt es momentan keine wirksame Therapie. Unser experimenteller Ansatz verwendet neueste Methoden der Neurowissenschaften, die eine Langzeitbeobachtung von strukturellen und funktionellen Netwerkveränderungen und Rückschluss auf die zellulären Grundlagen der Motorik erlauben. Ziel der Untersuchungen ist die Charakterisierung zentraler Ursachen von Motordefiziten, wie z.B. spastischem Muskeltonus, und die Entwicklung neuer neuromodulatorischer und regenerativer Verfahren zur gezielten Verbesserung der Motorik.
AG Neuroimaging and neuroengineering of experimental stroke
One third of stroke patients suffer from long-term disability and recovery of function is generally inclompete. By combining cutting-edge neurotechniques, such as in vivo MRI, viral tracing and light sheet microscopy, we seek to unravel the cellular and neural circuit mechanisms that underlie motor recovery after experimental stroke. Our studies dissect the development of motor deficits such as spastic muscle tone and develop novel neuromodulation and regeneration paradigms to enhance endogenous plasticity mechanisms and boost stroke recovery.
Dr. Philipp Boehm-Sturm (Charité Berlin, Experimental Neurology), Prof. Dr. Michael Hanke (Forschungszentrum Jülich, INM-7), Prof. Maged Goubran (University of Toronto, Dept. of Medical Biophysics, Toronto, Canada), Dr. Giovanna Ielacqua (Max-Delbrück-Centrum für Molekulare Medizin Berlin, Germany), Prof. Milos Pekny and Prof. Marcela Pekna (University of Gothenburg, Dept. of Clinical Neurosciences), Prof. Dr. Dr. Adele Rüger (Uniklinik Köln, Neurologie), Prof. Dr. Dr. Anna-Sophia Wahl (Institute for Stroke and Dementia Research, LMU Klinikum and Institute of Anatomy, Munich), Dr. Dirk Wiedermann (Max-Planck-Institut für Stoffwechselforschung, In-vivo-NMR)
Projects
Loss of function after stroke is caused by cell death and breakdown of the functional networks in the infarcted and connected brain regions. To some extent, the affected limb regains function, in a form of spontaneous recovery during the first 3 months after stroke. However, this recovery is generally incomplete and the underlying neural and molecular mechanisms are still unknown. In this project we are investigating brain/circuit activation patterns and analyse the transcriptome in the ipsi- and contralesional hemispheres of recovered vs. non-recovered mice after experimental stroke.
Publications:
Temporal and spatial gene expression profile of stroke recovery genes in mice.
Götz J, Wieters F, Fritz V, Käsgen O, Kalantari A, Fink GR, Aswendt M. Genes. 2023 Feb 9;14(2):454.
Lesion size- and location-dependent recruitment of contralesional thalamus and motor cortex facilitates recovery after stroke in mice.
Aswendt M*, Pallast N*, Wieters F*, Baues M, Hoehn M, Fink GR. Transl. Stroke Res. (IF 5.9) 2020 Mar 12. Epub ahead of print.
RNA-Sequencing Analysis Revealed a Distinct Motor Cortex Transcriptome in Spontaneously Recovered Mice After Stroke.
Ito, M., Aswendt, M., Lee, A.G., Ishizaka, S., Wang, E.H., Levy, S.L., Smerin, D., Leuze, C., Goubran, M., Cheng, M.Y., Steinberg, G.K. Stroke (IF 6.2) 2018; 49:2191-2199.
Optogenetic approaches to target specific neural circuits in post-stroke recovery.
Cheng, M., Aswendt, M., Steinberg G.K. Neurotherapeutics 13.2 (2016): 325-340.Conference abstracts:319.05 / U13 - Investigating neural and molecular mechanisms of spontaneous recovery in experimental stroke. Ito, M., Aswendt, M., Cheng, M., Ishizaka, S., Lee, A., Levy, S., Smerin, D., Wang, E., Steinberg, G.K. Neuroscience 2016, San Diego, USA.
Working with in vivo and ex vivo imaging at different spatial resolutions requires a common atlas registration for cross-modality comparisons. For example, if we detect connectivity change in primary motor cortex with MRI, it will be of interest to investigate the same brain area with histology. In order to merge in vivo MRI with 2D histology, we have created software pipelines for automated processing of structural and functional mouse brain MRI (AIDAmri) and automated cell counting - optimised for mouse brain histology and immunostainings (AIDAhisto). These pipelines include accurate approaches to register whole brain (MRI) and single slices (histology) respectively with the Allen Mouse Brain Atlas.
The software is available on Github. Chat with us and find answers to FAQs on Gitter.
Publications:
Network analysis of neuroimaging in mice. Scharwächter L, Schmitt F, Pallast N, Fink GR, Aswendt M. (2022) Neuroimage (253).
Atlas-based imaging data analysis tool for quantitative mouse brain histology (AIDAhisto). Pallast N. Wieters F, Fink GR, Aswendt M. (2019) Journal of Neuroscience Methods (326).
Processing Pipeline for Atlas-based Imaging Data Analysis of Structural and Functional Mouse Brain MRI (AIDAmri). Pallast N, Diedenhofen M, Blaschke S, Wieters F, Wiedermann D, Hoehn M, Fink GR, Aswendt M. (2019). Frontiers in Neuroinformatics, 04 June 2019.
Whole-Brain Microscopy Meets In Vivo Neuroimaging: Techniques, Benefits, and Limitations. Aswendt, M., Schwarz, M., Abdelmoula, W. M., Dijkstra, J., & Dedeurwaerdere, S. (2016). Molecular Imaging and Biology, 1-9.
To obtain valid scientific results in pre-clinical research, standardization of experimental protocols and data handling need to be set before starting the actual experiment. Efficient data management becomes more and more important with the increasing number and variety of experimental procedures. However, to date, preclinical research is still in a transition phase. While the recorded data type is predominantly electronic data, the documentation is still the lab notebook. We designed a database template which is useful for all labs working with laboratory animals and the adaption to specific research projects requires no prior scripting expertise. The database works operating-system independent through the web browser and allows multiple user to work simultaneously. The data entry is monitored and restricted for particular tests according to the user management in order to keep for example users during the experiment blinded for the experimental group. Furthermore, we have established an efficient workflow to store and share the experimental data according to FAIR prinicples.
Video Database Tutorial: Supplementary Data
The software and manual are available on GitHub. Need help? Chat with us and find answers to FAQs in the AIDA_Tools Gitter rooms.
Publications:
How to establish and maintain a multimodal animal MRI data repository using Datalad.
Kalantari A, Aswendt M, Hanke M, Szczepanik M, Heunis S, Mönch C, Wachtler T. Sci. Data. 2023 Jun 5;10:357.
Cloud-based relational database for multimodal animal data.
Pallast, N., Wieters F., Nill M., Fink G.R., Aswendt M.
Oxford Database 2018
Resources:
Link to download the research database template. To open the template, a Ninox www.ninoxdb.de account is required.
Spasticity is a common symptom after stroke, arising in about 30% of patients, and affecting the lower as well as upper limb. The spastic (abnormal) muscle tone appears in the first weeks after stroke onset and maladaptive processes lead to a manifestation of spasticity, which ultimately limits the success of rehabilitation. While previous studies investigated the abnormal intra-spinal processing, spasticity remains a result of damage to the primary motor regions. Therefore we are characterizing the cellular components and network changes which lead to spasticity after stroke to effectively develop opposing optogenetic and chemogentic stimulation paradigms.
Publications:
Terminal H-reflex measurements in mice. Wieters F, Gruhn M, Büschges A, Fink GR, Aswendt M. J Vis Exp. 2022 In press
Introduction to spasticity and related mouse models. Wieters F, Weiss Lucas C, Gruhn Matthias, Büschges A, Fink GR, Aswendt M. Exp Neurol. 2021 Jan;335:113491.
How can we quantify the fate of cultured or transplanted stem cells noninvasively? This fundamental question arises from our previous studies on stem cell-induced recovery processes in experimental stroke. We follow transplanted stem cells longitudinally upon transplantation into the mouse brain by noninvasive imaging techniques. However, while cell localization is thus possible, survival and differentiation of the graft cannot be detected. Yet, there is a great need to gain insight into graft-host reactions and the biological time profile of stem cell actions before stem cell therapy can be further translated. Approaches with conventional techniques reveal the cell status only upon sacrificing the animal and cannot be performed on the same sample, which prohibits the intra-individual time profile. We take a different approach towards quantitative monitoring of stem cell viability and differentiation for both, in vitro and in vivo conditions using bioluminescence reporters.
Publications:
The in vivo timeline of human neural stem cell differentiation.
Vogel S, Schäfer C, Hess S, Minassian A, Schwarz M, Folz-Donahue K, Kukat C, Ehrlich M, Zaehres H, Kloppenburg P, Hoehn M, Aswendt M. Stem Cell Research (IF 3.9), Volume 37, May 2019, 101429.
Neurogenesis upregulation on the healthy hemisphere after stroke enhances compensation for age-dependent decrease of basal neurogenesis.
Adamczak J.*, Aswendt M.*, Kreutzer C., Rotheneichner P., Riou A., Selt M., Beyrau A., Uhlenküken U., Diedenhofen M., Nelles M., Aigner L., Couillard-Despres S., Hoehn M. (2017). Neurobiology of Disease, 99, 47-57.
Human neural stem cell intracerebral grafts show spontaneous early neuronal differentiation after several weeks.
Tennstaedt, A., Aswendt, M., Adamczak, J., Collienne, U., Selt, M., Schneider, G., ... & Kloppenburg, P. (2015). Biomaterials, 44, 143-154.
A multi-modality platform to image stem cell graft survival in the naïve and stroke-damaged mouse brain.
Boehm-Sturm, P., Aswendt, M., Minassian, A., Michalk, S., Mengler, L., Adamczak, J., ... & Hoehn, M. (2014). Biomaterials, 35(7), 2218-2226.
Boosting bioluminescence neuroimaging: an optimized protocol for brain studies
Aswendt, M., Adamczak, J., Couillard-Despres, S., & Hoehn, M. (2013). PloS one, 8(2), e55662.
Evaluating reporter genes of different luciferases for optimized in vivo bioluminescence imaging of transplanted neural stem cells in the brain
Mezzanotte, L., Aswendt, M., Tennstaedt, A., Hoeben, R., Hoehn, M., & Löwik, C. (2013). Contrast media & molecular imaging, 8(6), 505-513.
Das Team
Aref Kalantari, Doktorand
Fatemeh Mahani, Doktorandin
Victor Vera Frazao, Wissenschaftlicher Mitarbeiter
Carolin Hambrock, Masterarbeit
Thomas Kühnel, Wissenschaftliche Hilfskraft
Marko Schmelzer, Bachelorarbeit
Marc Schneider, Wissenschaftliche Hilfskraft
Jule Ullert, Bachelorarbeit
Elizabete Vetra, Studentische Hilfskraft
Franziska Ziolkowski, Masterarbeit
Labor:
Haus 74 (FLEX)
Uniklinik Köln
Weyertal 115C
50931 Köln
Büro:
Haus 27 (Kapellchen), 1. Etage, Raum 001/2
Kerpener Str. 62
50937 Köln
Telefon + 49 221 478-86416 und -30626
Telefax +49 221 478-30808
Für interessierte Studierende der Biologie, Neurowissenschaften, Physik und Informatik bieten wir jederzeit Bachelor- oder Masterarbeiten an! Bewerbung mit Kurzlebenslauf und aktuellen Transcript of Records per E-Mail an Dr. Markus Aswendt.